[ad_1]
Construction-guided linker design
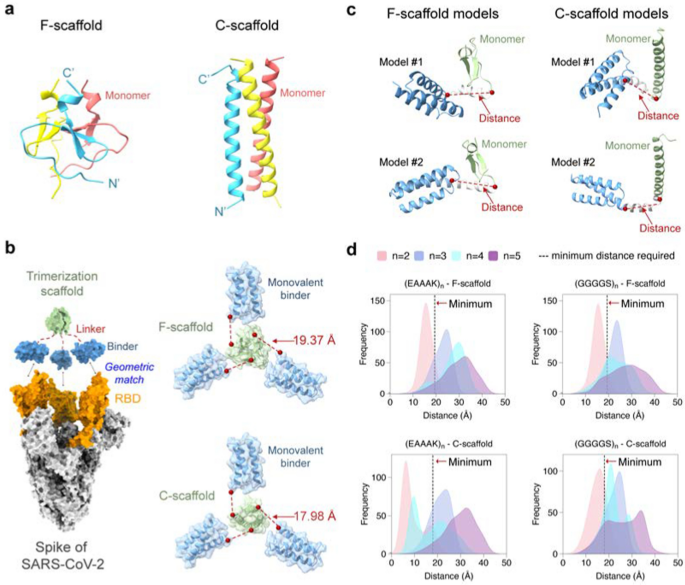
To design a self-assembling trivalent protein with desired physiochemical properties and therapeutic efficacy, we rigorously chosen two important parts: the monovalent therapeutic agent and the trimerization scaffold. As talked about above, microproteins are effectively suited to be engineered into multivalent codecs as a result of their small measurement, stability, and ease of manufacturing. As a proof of idea, we first used the microprotein LCB3 [26] as a monovalent binder to the S protein of SARS-CoV-2. This microprotein (MP) is a mini-mimetic of the ACE2 protein with solely 64 aa (hereafter we seek advice from it as miniACE2) and has been reported to bind to the RBD of the S protein. To acquire optimum multivalent constructs, we then chosen two well-studied self-assembling domains because the take a look at trimerization scaffolds, specifically the β-propeller-like foldon area of T4 fibritin utilized in vaccines [27,28,29] and an α-helical coiled-coil peptide [30], which shall be known as F-scaffold and C-scaffold, respectively (Fig. 2a).
Construction-guided linker design. a Two trimerization scaffolds used on this examine. b The left panel: The trivalent assemble designed to match the geometry of the three binding websites on RBDs (orange) of the S protein (silver). The monovalent binder and the trimerization scaffold are proven in blue and inexperienced, respectively. The precise panel: The calculated minimal distances required between the binder and scaffold. c Schematic of linker fashions generated by RosettaRemodel. For every mannequin, the gap from the C-terminus of the binder to the N-terminus of the scaffold was decided. d Distributions of the talked about distances of the designed linkers of various lengths. The dotted black traces point out the minimal distances required for the constructs
We hypothesized {that a} well-designed trivalent protein may concurrently have interaction all three RBDs of the S protein, thereby blocking the ACE2 binding and enhancing its neutralizing exercise towards the SARS-CoV-2 variants. It has been proven that RBD can undertake two totally different conformations: standing-up conformation (RBD-up) for receptor binding and lying-down conformation (RBD-down) for immune evasion [7, 31, 32]. The RBD-up state is important for membrane fusion and virus entry [9, 31], and doubtlessly facilitates immune clearance. Subsequently, our designed objective was to lure the lively RBD-up conformation by totally occupying all three RBDs with a designed trivalent protein. To attain this, we superimposed the miniACE2-RBD complicated (PDB ID: 7JZM) onto the S protein with the 3-RBD-up state (PDB ID: 7CT5). Primarily based on the superimposed construction, we calculated the minimal distance required for a linker to attach miniACE2 and the given trimerization scaffold utilizing the Lagrange multiplier methodology, and located that the minimal distances for the F- and C-scaffolds are 19.37 Å and 17.98 Å, respectively.
Primarily based on the minimal distances, we then designed linkers to attach the monovalent binder and the trimerization scaffold, guaranteeing an acceptable geometry to match the homo-trimeric goal websites. We chosen two broadly used penta-peptide fragments, the versatile GGGGS and the inflexible EAAAK [33], because the constructing fragment for the candidate linkers, after which decided the repeat quantity (n) of the given fragment within the linker in accordance with the minimal distances and the folding conformations of every linker. Contemplating the utmost size of the prolonged conformation of a penta-peptide (GGGGS or EAAAK), no less than two copies of the fragments (i.e., 10-aa size) are wanted for a linker to attach the binder to the scaffold. To find out the optimum repeat quantity n, we used RosettaRemodel to pattern a lot of the lowest-energy folding conformations for the linkers of (GGGGS)n or (EAAAK)n (n = 2, 3, 4, or 5) (see Supplies and strategies). For every n, the folding conformations of the given linker had been predicted with the binder at its N-terminus and the scaffold on the C-terminus; and within the calculations, each the binder and the scaffold had been handled as inflexible our bodies. After the conformational sampling, the 1000 top-ranking lowest-energy conformations had been used to calculate the distributions of the distances between the C-terminus of the binder and the N-terminus of the scaffold (Fig. 2).
As proven in Fig. 2b, many of the sampling distances for the linkers (GGGGS)2 and (EAAAK)2 fell wanting the above-mentioned minimal distances required for the geometric matching of the binder and the trimerization scaffold, indicating that linkers designed with n = 2 might not be appropriate. In distinction, most distances for the linkers (GGGGS)n and (EAAAK)n, with n = 3, 4, or 5, exceeded the minimal distances, indicating that n ≥ 3 is required. Amongst these, the gap distributions for n = 3 had been comparatively slim, whereas these for n = 4 or 5 had been wider, indicating that extra conformations had been energetically doable for these linkers and thus that the binders related to the trimerization unit may bind to extra positions in area, permitting them to adapt their conformations to totally different epitopes on the goal protein. Nonetheless, the gap distributions for n = 4 had been irregular, neither as slim as n = 3 nor as broad as n = 5; moreover, these for n = 3 and 5 have coated many of the vary seen in n = 4, making n = 4 much less preferable. Additionally, provided that n = 5 already coated the doable distances, we now not explored the scenario of n > 5.
Lastly, we selected 4 take a look at linkers, the versatile (GGGGS)3 and (GGGGS)5, and the inflexible (EAAAK)3 and (EAAAK)5, to assemble the trivalent proteins for miniACE2, leading to eight trivalent proteins tailor-made for the 2 trimerization scaffolds. We designated the 2 proteins utilizing versatile (GGGGS)n and C-scaffold as MP-3fc, MP-5fc, respectively; these utilizing (GGGGS)n and F-scaffold as MP-3ff, MP-5ff, respectively; these utilizing inflexible (EAAAK)n and C-scaffold as MP-3rc, MP-5c, respectively; and people utilizing (EAAAK)n and F-scaffold as MP-3rf, MP-5rf, respectively (Extra file 1: Desk S1).
Molecular simulation analysis of trivalent constructs
Biomedical and therapeutic purposes of multivalent proteins often require them to have good physicochemical properties corresponding to environment friendly self-assembly and good conformational homogeneity. To determine the very best candidate among the many eight constructs, we evaluated these properties of the eight constructs utilizing molecular dynamics (MD) simulations. For every assemble, three unbiased simulations had been carried out, every with a simulation time of 300 ns. Then, we calculated the basis imply sq. deviation (RMSD) of the protein spine heavy atoms throughout the simulation trajectories, utilizing their preliminary constructions because the reference conformations (Extra file 1: Fig. S1). As could be seen in (Extra file 1: Fig. S1, the RMSD outcomes confirmed that every one the programs reached equilibrium after about 150-ns simulations. Subsequently, we used the post-150 ns trajectories for the next analyses.
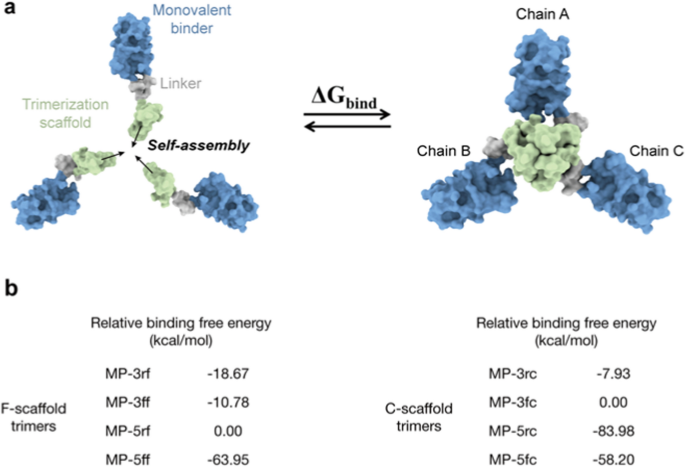
To evaluate the self-assembly talents, we first used the Molecular Mechanics/Generalized Born Floor Space (MM/GBSA) methodology to estimate the binding free energies between the three monomers of the trivalent constructs, as illustrated in Fig. 3a and (Extra file 1: Desk. S2). Though MM/GBSA has limitations in predicting absolute values of binding free vitality, it excels in rating the relative binding affinities of various molecules [34]. Equally, the relative binding free energies of various constructs may additionally rank their self-assembly talents. The MM/GBSA calculations confirmed that the binding free energies of the F-scaffold constructs are sometimes decrease than these of the C-scaffold constructs, aside from that of MP-5rc (Extra file 1: Desk S2). This implies that the self-assembly talents of F-scaffold constructs are comparatively higher than these of the C-scaffold constructs. Among the many F-scaffold constructs, the binding free vitality of MP-5rf is the best; as proven in Fig. 3b, the relative binding free energies of the opposite three constructs are unfavourable, indicating that their self-assembly talents are stronger than MP-5rf. Of them, MP-5ff has the bottom relative binding free vitality, implying the strongest self-assembly tendency. For the C-scaffold constructs, MP-5rc has the bottom relative binding free vitality, indicating a stronger self-assembly skill, particularly in contrast with the upper relative vitality values of MP-3fc and MP-3rc.
Molecular simulation evaluation of the trimerization tendency of trivalent constructs. a Schematic diagram for the binding course of to calculate the binding free vitality of a given trivalent assemble. b Relative binding free energies of the trivalent constructs calculated by MM/GBSA. For F-scaffold trimers, MP-5rf was used because the reference to calculate the relative values of different constructs. For C-scaffold trimers, the reference is MP-3rc. Please see the MM/GBSA uncooked knowledge in (Extra file 1: Desk S2
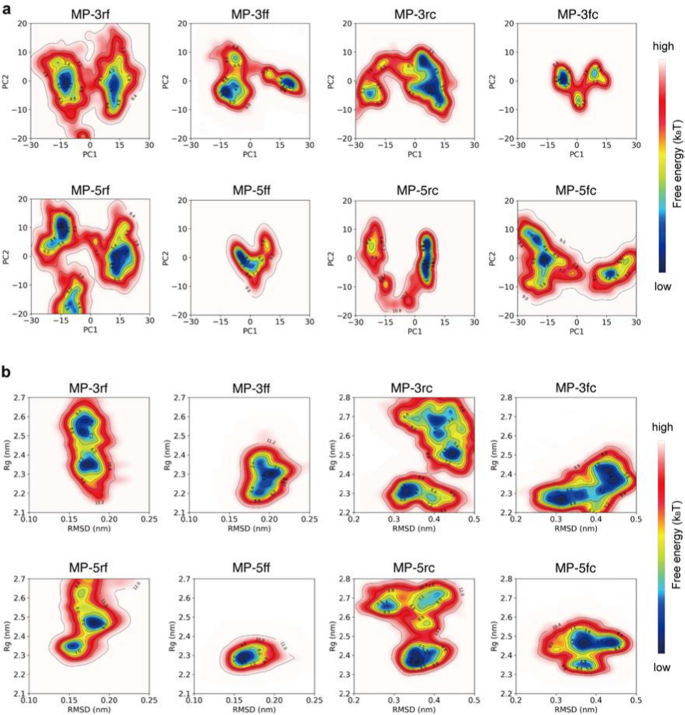
To analyze the conformational homogeneity, we first carried out principal element evaluation (PCA) on the simulation trajectories after which mapped the simulated conformations of the proteins onto the ensuing principal parts to generate their free vitality landscapes (FELs) (see Strategies and supplies). For example, the PCA outcomes for a trajectory of MP-5ff are proven in (Extra file 1: Fig. S2). As seen, the primary two principal parts contributed to over 80% of the cumulative variance and had been thus thought-about PC1 and PC2 (Extra file 1: Figs. S2A, D). By projecting the simulated conformations onto the two-dimensional area of PC1 and PC2, the ensuing FELs confirmed the distribution patterns of the simulated constructs and their doable numbers of dominant trimer-like conformations within the simulations (Fig. 4).
Free vitality landscapes (FELs) for the MD conformations of the trivalent constructs. a FELs of conformational projections onto the primary and the second principal parts (PC1 and PC2). b FELs of conformational projections onto two different response coordinates: root imply sq. deviation (RMSD) and radius of gyration (Rg)
As proven in Fig. 4a, the FEL patterns of the eight constructs aren’t an identical, with about 1–3 low-energy wells (in blue) indicating totally different numbers of dominant trimer-like conformations within the simulations. Considerably, aside from MP-5ff, different constructs had a wider or a couple of low-energy effectively, corresponding to MP-5rc with a wider low-energy effectively, MP-3rf, MP-5rf with 2 low-energy wells, and MP-3ff, MP-3fc, MP-5fc with 3 low-energy wells. Thus, solely MP-5ff confirmed solely a low-energy trimer conformation within the simulations, suggesting that the conformational homogeneity of this protein is the very best.
To additional affirm the FEL outcomes, we additionally assemble FELs by mapping the simulated conformations onto the two-dimensional area outlined by two different response coordinates: root imply sq. deviation (RMSD) and radius of gyration (Rg) (Fig. 4b). In keeping with the outcomes of Fig. 4a, besides MP-5ff the FELs of all constructs had a number of low-energy wells (in blue), suggesting that a number of low-energy trimer-like conformations coexist within the simulations. Taken collectively, the ends in Fig. 4 prompt that MP-5ff doubtless has a single secure trimer conformation and thus the very best conformational homogeneity.
Experimental validation and practical take a look at
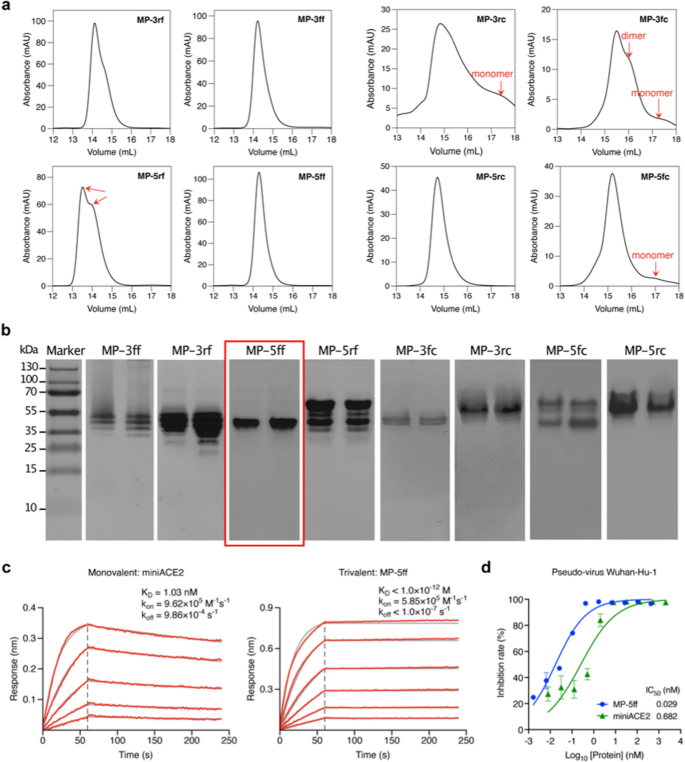
To validate the computational outcomes, we expressed the 8 designed constructs in E. coli Rosetta (DE3) cells and purified the proteins utilizing Ni–NTA affinity chromatography. We then characterised their oligomeric states in resolution by size-exclusion chromatography (SEC). As proven in Fig. 5a, the SEC profiles revealed that the 4 F-scaffold proteins had narrower and sharper trimer peaks than the C-scaffold proteins, indicating a better trimer ratio within the F-scaffold constructs. Among the many F-scaffolds, the height of MP-5ff seems to be the sharpest and probably the most concentrated one, indicating that it’s the best trivalent assemble, in good settlement with the computational analysis. In distinction, MP-5rf displayed two distinct peaks, in all probability comparable to the specified trimerization conformation and one other oligomeric state. Certainly, the binding free vitality calculations in Fig. 3b have already indicated that the MP-5rf trivalent assemble is much less secure than the opposite three F-scaffold constructs. As for the C-scaffold constructs, solely MP-5rc had a pointy, single peak indicating a trimer; nonetheless, apart from the trimer peak, the opposite three constructs had detectable monomer or dimer peaks, particularly MP-3rc and MP-3fc, suggesting that that they had a decrease trimer ratio. Clearly, these outcomes confirmed the MM/GBSA calculations (Fig. 3b) and confirmed that these constructs with the decrease binding free energies have greater trimerization efficiencies.
Experimental verifications of the trivalent constructs of miniACE2. a SEC profiles of the purified proteins of the constructs. b Native-PAGE evaluation of the protein trimer fractions remoted from SEC. c BLI measurements of the binding kinetics of the monovalent miniACE2 and the trivalent assemble, MP-5ff, to the immobilized RBD of SARS-CoV-2 (Wuhan-Hu-1). Purple traces characterize the uncooked knowledge and the kinetic suits are proven in grey. d Neutralizing exercise of miniACE2 and MP-5ff towards SARS-CoV-2 pseudovirus (Wuhan-Hu-1)
The FEL analyses within the final subsection have prompt that even within the trimer state, the investigated trivalent constructs are prone to include a number of totally different trimer-like conformations (Fig. 4). To additional examine the doable distributions of trimerization conformations, we carried out Native-PAGE evaluation on the protein samples collected from the SEC trimer peaks, as a result of this method can separate two or extra trimer-like conformations of the trivalent proteins. As proven in Fig. 5b, among the many eight constructs, solely MP-5ff introduced a single protein band, indicating a single secure trimer conformation, which is in keeping with the computational prediction displaying solely a single vitality effectively within the FELs (Figs. 4a, b). In distinction, MP-3ff and MP-3rf confirmed a number of distinct bands, indicating that they will undertake a number of coexisting conformations. For MP-5rf, we noticed two dominant bands with a number of fainter ones at numerous positions; the higher one might recommend the formation of bigger oligomers that fail to take care of a secure trimer. Equally, MP-5fc additionally displayed such a sample. For the opposite three C-scaffold constructs, we additionally noticed a couple of band: MP-3fc exhibited two clear bands, whereas MP-3rc and MP-5rc confirmed a transparent one and a number of other fainter bands. These findings validate the computational predictions that a number of low-energy trimer-like conformations might coexist for these constructs (Figs. 4a, b). Because of this, MP-5ff was discovered to be the very best assemble with the best trimerization effectivity and conformational homogeneity.
We subsequent examined the goal binding affinity of the optimum assemble, MP-5ff, to RBD of the S protein utilizing Biolayer interferometry (BLI). Since miniACE2 was initially designed to particularly goal the SARS-CoV-2 Wuhan-Hu-1 pressure [26], right here we centered our practical analysis on miniACE2 and MP-5ff towards this particular pressure. As proven in Fig. 5c, MP-5ff exhibited a a lot slower dissociation fee (okayoff < 1.0 × 10–7 s−1) in comparison with that of miniACE2 (okayoff = 9.86 × 10–4 s−1); thus, the ensuing equilibrium dissociation fixed (OkayD) is lower than 1 pM, whereas that of miniACE2 is 1.03 nM. Thus, the binding affinity of MP-5ff for RBD is 1000-fold higher than that of its monovalent counterpart miniACE2, clearly demonstrating that protein multivalency may considerably improve the goal binding affinity. Then, we evaluated the neutralizing actions of miniACE2 and MP-5ff towards SARS-CoV-2 pseudovirus (Wuhan-Hu-1). As indicated in Fig. 5d, the monovalent miniACE2 was already in a position to inhibit the virus with an IC50 of 682 pM; nonetheless, the trivalent MP-5ff nonetheless considerably enhanced the neutralizing exercise (IC50 = 29 pM), exhibiting a 23-fold enhance. Taken collectively, the trivalent MP-5ff designed by our rational method has glorious physicochemical properties and potent antiviral exercise.
Engineering of a broad-spectrum trivalent nanobody
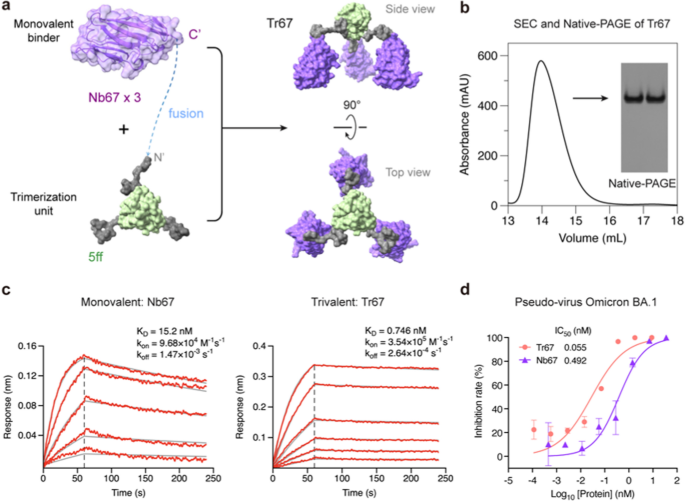
To additional exhibit the effectiveness of our method, we utilized the 5ff trimerization unit to engineer a trivalent nanobody focusing on the dominant circulating Omicron variants, as a result of nanobodies characterize one other broadly used class of microproteins effectively fitted to multivalent building. For this function, we chosen Nb67, a nanobody recognized by Xiang et al. [35] from serially immunized camelid sera, which was reported to neutralize Omicron BA.1. By fusing Nb67 with the 5ff trimerization unit, we created a trivalent nanobody Tr67 (Fig. 6a, (Extra file 1: Desk S1). Following the identical computational and experimental procedures efficiently employed for MP-5ff, we assessed the trimerization effectivity and conformational homogeneity of the engineered Tr67 utilizing MD simulations ((Extra file 1: Fig. S3), SEC and native-PAGE analyses (Fig. 6b). These obtained outcomes demonstrated that Tr67 has a trimerization effectivity and conformational homogeneity similar to that of MP-5ff.
Design and experimental characterization of Tr67. a Schematic diagram illustrating the trimerization of Nb67 nanobody fused with the optimum trimerization unit 5ff (see its amino-acid sequence of the fusion monomer of Nb67 with 5ff in (Extra file 1: Desk S1). b SEC and Native-PAGE evaluation of Tr67, displaying excessive trimerization tendency and conformational homogeneity. c BLI measurement of the binding kinetics of the monovalent Nb67 and Tr67 to the immobilized RBD of SARS-CoV-2 pseudovirus (Omicron BA.1). d Neutralizing actions of Nb67 and Tr67 towards SARS-CoV-2 pseudovirus (Omicron BA.1). Three unbiased experiments had been carried out
We then measured the binding affinity of Tr67 to the goal RBD of the S protein utilizing BLI and its neutralizing exercise towards SARS-CoV-2 pseudoviruses. As proven in Fig. 6c, Tr67 exhibited a better affiliation fee (okayon) and a decrease dissociation fee (okayoff) in comparison with its monovalent counterpart Nb67. The ensuing OkayD was 0.746 nM, which is an about 20-fold enhance in affinity in comparison with that of the monovalent Nb67 (OkayD = 15.2 nM). Equally, Tr67 confirmed a lot stronger inhibitory exercise towards the SARS-CoV-2 Omicron BA.1 pseudovirus than Nb67, with an IC50 of 55 pM versus 492 pM for Nb67 (Fig. 6d). These outcomes demonstrated that the trivalent nanobody has an enhanced efficiency and thus higher potential to fight viral infections in comparison with its monovalent counterpart.
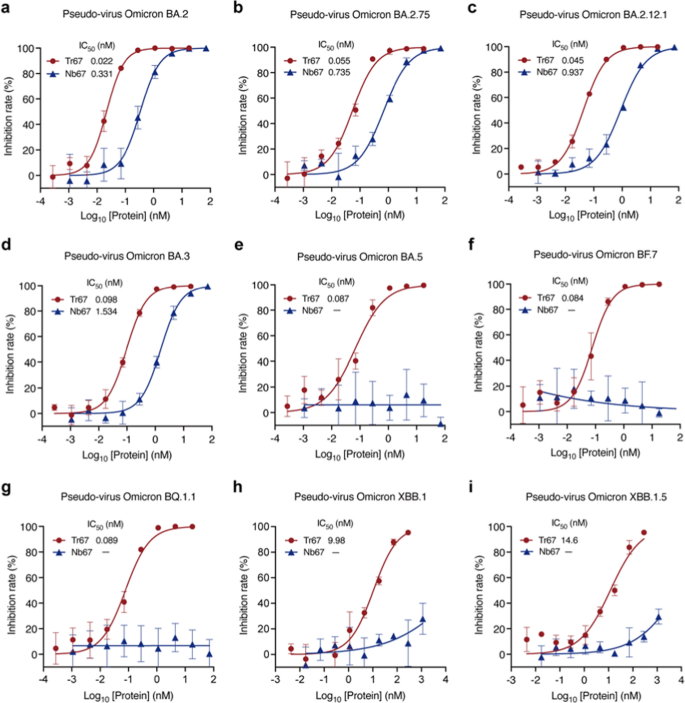
To additional examine the broad-spectrum neutralizing potential of Tr67, we evaluated its neutralization actions towards the dominant Omicron variants (Fig. 7). For Omicron BA.2, Tr67 exhibited an IC50 of 0.022 nM and that of Nb67 is 0.331 nM, so the neutralizing exercise was tremendously enhanced by about 15 folds (Fig. 7a). Comparable enhancements had been noticed for Omicron BA.2.75, BA.2.12.1, and BA.3: the corresponding IC50 values of Tr67 had been 0.055, 0.045, and 0.098 nM, respectively, and people of Nb67 had been 0.735, 0.937, and 1.534 nM, respectively (Figs. 7b–d). Unexpectedly, Tr67 additionally neutralized the variants which might be extra prone to evade humoral immunity. For Omicron BA.5, BF.7, and BQ.1.1, Nb67 failed to attain any detectable neutralization; nonetheless, Tr67 neutralized them with IC50 values of 0.087, 0.084, and 0.089 nM, respectively (Figs.7e–g). Even for probably the most immune-evasive Omicron XBB household, Tr67 nonetheless maintained neutralizing exercise, however Nb67 didn’t (Figs.7h, i). Particularly, the IC50 values of Tr67 towards XBB.1 and XBB.1.5 had been 9.98 and 14.6 nM, respectively. Thus, in contrast with its monovalent counterpart, Tr67 has a major enhance within the neutralizing exercise towards all of the examined Omicron variants, suggesting that multivalent proteins have the potential to be developed into broad-spectrum medication towards the rising SARS-CoV-2 variants.
Broad-spectrum neutralization potential of Tr67 towards the dominant SARS-CoV-2 Omicron variants. a—i Neutralizing actions towards SARS-CoV-2 pseudoviruses Omicron BA.2, BA.2.75, BA.2.12.1, BA.3, BA.5, BF.7, BQ.1.1, XBB.1, and XBB.1.5 variants, respectively. Three unbiased experiments had been carried out for every variant
Cryo-EM evaluation of Tr67-spike complicated
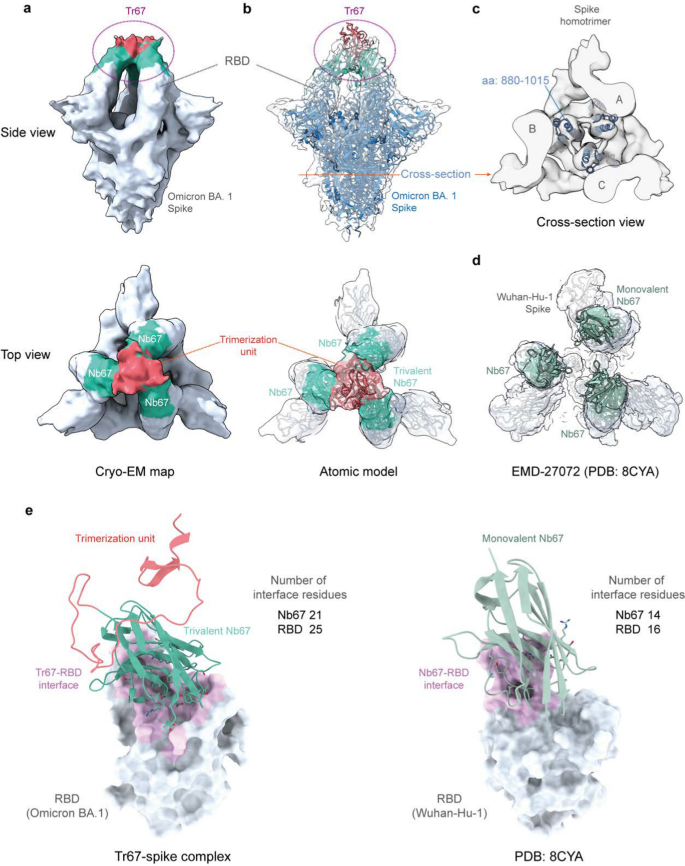
Lastly, to verify whether or not the binding mode of Tr67 to the spike protein is in keeping with our design, we decided the complicated construction of Tr67 with the Omicron BA.1 spike protein utilizing cryo-EM ((Extra file 1: Figs. S4, Desk S3 and Fig. 8). As seen in Fig. 8A, the cryo-EM density map obtained by the single-particle 3D reconstruction methodology clearly reveals that the complicated construction is a triple-symmetric homotrimer; furthermore, the density of Tr67 certain to the RBDs on the high of the S-protein could be very effectively outlined (Fig. 8a, aspect view), and the density of the three Nb67 nanobodies and the trimerization unit 5ff (Fig. 8a, high view) will also be distinguished. Subsequently, the cryo-EM construction supplied experimental proof that Tr67 is certainly certain to the epitopes specified by the computational design.
Cryo-EM constructions of Tr67 in complicated with the SARS-CoV-2 (Omicron BA.1) spike protein. a EM density map of Tr67-spike complicated in “3-RBD-up” conformation on the total decision of 9 Å. b Becoming of the atomic mannequin of the designed Tr67-spike complicated into the EM density map. The EM density is proven as a clear grey floor and the spike protein is rendered in blue. c Cross-section view of the stem area of the spike protein. d An open-like, “3-RBD-up” conformation with the SARS-CoV-2 (Wuhan-Hu-1) spike protein induced by three separated Nb67. e The binding interfaces of trivalent (left panel) and monovalent (proper panel) Nb67 binding with the RBDs (proven as grey floor) are proven in pink, and the contact residues on Nb67 are proven as sticks
To acquire the atomic mannequin of the cryo-EM construction, we used the MD versatile becoming methodology to suit the atomic mannequin of the Tr67-spike complicated constructed within the computational design into the density map (Fig. 8b). As could be seen, the atomic mannequin of the entire complicated fitted effectively into the map; and this turned clearer by the illustration of the central alpha-helical areas within the S protein stem (Fig. 8c). Extra particularly, the designed Tr67 matches effectively with corresponding densities, displaying that Tr67 precisely binds to the specified positions on the S protein (Fig. 8b, high view).
Considerably, we discovered that the complicated construction is one during which all 3 RBDs of the S protein are within the standing-up state (3-RBD-up). Because of its amino acid mutations and RBD-RBD interactions, the Omicron spike is often stabilized within the “2-RBD-down, 1-RBD-up” conformation; this conformational state was thought-about to facilitate the up-RBD to method ACE2 after which to advertise membrane fusion [36,37,38]. Not like the spikes of early variants corresponding to Wuhan-Hu-1, the Omicron spike hardly ever happens within the 3-RBD-up conformation, which can must be induced by a mixture of distinct antibodies [39, 40]. Thus, the cryo-EM construction did affirm that Tr67 can induce the Omicron spike into the 3-RBD-up conformation. Word that, in contrast to the widespread 3-RBD-up conformation (Wuhan-Hu-1) induced by monovalent nanobodies, during which the three RBDs are in an open-like, unassociated state (Fig. 8d), Tr67 has a further trimerization unit that covalently hyperlinks the three binders and thereby firmly locks the three RBDs in an inactive state (Fig. 8b, high view). Unsurprisingly, the S protein in such a Tr67-bound state can’t bind ACE2 anymore and due to this fact its membrane fusion operate is totally inhibited.
To grasp the molecular foundation of the elevated binding affinity of Tr67 for the S protein, we analyzed the binding interfaces of monovalent Nb67 and Tr67 with the RBDs utilizing the Nb67-spike construction and the atomic mannequin of the Tr67-spike complicated. We recognized the interface (contact) residues by a 4-Å distance cutoff between the atoms of Nb67 and people of the RBD, as proven in Fig. 8e and listed in (Extra file 1: Desk S4. As could be seen, the binding websites of Nb67 and Tr67 on the RBDs are an identical to these of ACE2. Nonetheless, the Nb67 binder in Tr67 has a bigger contact space with the Omicron RBD, and the interface comprises 21 residues from Nb67 and 25 residues from the Omicron RBD. In distinction, the monovalent Nb67 and the Wuhan Hu-1 RBD have solely 14 and 16 interfacial residues, respectively. As well as, the Nb67 binder in Tr67 varieties extra hydrogen bonds and salt bridges (Extra file 1: Desk S4), suggesting stronger binding interactions. In keeping with the BLI ends in Fig. 6c, the variety of interfacial residues additionally helps that Tr67 may set up a extra intensive community of interactions, contributing to stronger binding to the Omicron BA.1 and thus enhancing its neutralizing exercise.
To elucidate why Tr67 additionally binds to different Omicron variants (Extra file 1: Fig. S5), we constructed structural fashions for his or her RBDs based mostly on the atomic fashions in Fig. 8b, after which analyzed their binding interfaces with the monovalent Nb67 and Tr67 by docking simulation utilizing PyDock [41]. The perfect-scoring binding poses from the simulations had been used because the representatives of the Nb67-spike and Tr67-spike complexes, as proven in (Extra file 1: Figs. S6 and S7, respectively. As illustrated in (Extra file 1: Fig. S6a, for the variants BA.1, BA.2, BA.2.75, BA.2.12.1, and BA.3 (cluster 1), Nb67 was efficiently docked into the anticipated epitope on RBD; nonetheless, for variants BA.5, BF.7, BQ.1.1, XBB.1, and XBB.1.5 variants (cluster 2), Nb67 within the best-scoring poses was not situated on the anticipated websites, however at different websites which might be sterically unfavorable within the 1-RBD-up conformation of the S protein (Extra file 1: Fig. S6b). In keeping with this, Nb67 was in a position to neutralize the Omicron variants in cluster 1 however not these in cluster 2 (Fig. 7). Clearly, the amino-acid mutations of the cluster 2 variants weaken the interactions of the monovalent Nb67 with the variant RBD websites in Fig. 8e and thus abolish the neutralization. Notably, just like a earlier examine [35], we discovered that the mutation at 486 performs a key position on this course of (Extra file 1: Fig. S5 and Desk S5). In distinction, for all variants, Tr67 of the best-scoring poses binds to the identical epitopes as that of BA.1. The binding interfaces are additionally bigger than these of Nb67 (Extra file 1: Desk S6). It seems that the synergistic binding of the three Nb67s in Tr67 will increase the binding interface after which results in greater binding affinities that might resist the mutations corresponding to that at 486 to some extent. Because of this, Tr67 remains to be in a position to bind to the identical epitopes of the Omicron BA.1 S protein and neutralize the opposite variants examined. Clearly, additional structural research are wanted to elucidate the detailed molecular mechanisms concerned.
[ad_2]